Molecular Biology and Genomics: Key Concepts and Advanced Techniques
Document about Molecular Biology and Genomics. The Pdf, a detailed set of notes for University-level Biology students, explores key concepts and advanced techniques in molecular biology and genomics, including PCR, FANTOM, and RNA sequencing.
See more25 Pages

Unlock the full PDF for free
Sign up to get full access to the document and start transforming it with AI.
Preview
Introduction to Genomics
Genomics is defined as the study of genes and their functions (with related techniques). It involves: · mapping · sequencing · functional analysis of genomics -> HOW IS GENOMICS DIFFERENT FROM GENETICS? Genomics was first used to identify collections of genes, while nowadays the term includes all the DNA.
4 GENOMICS -+ TRANSCRIPTOMICS 1 the -omies analyze +- PROTEOMICS the large scale of collections of genes 1 genetics analyzes the functioning and composition of ONE SINGLE gene 1 genomics is addressed to all genes, their function and their inter-relationship, to identify their combined influence on the growth and development of the organism Genomics became came on independent field after the Human Genome Project (HGP) 1 development of genomics : · Recombinant DNA technology · HGP BASIC . Bioinformatic
Recombinant DNA Technologies
Polymerase Chain Reaction (PCR)
PCR (polymerase chain reaction) allows selection of a SPECIFIC DNA SEQUENCE. The process was possible only thanks to the isolation and biochemical characterization enzymes (from bacteria), in order for them to be used in vitro. - > restriction enzymes at With PCR it was possible to amplify in vitro specific DNA of specific points DNA fragments (in this way insufficiency in DNA quality was no longer a limitation). The mechanism behind PCR consists in the capacity of DNA to be converted from its double helix form to the single stranded form by heat changes. By raising the temperature the DNA is denatured, by lowering it, the DNA hybridizes back to its double stranded form. However, we need enzymes able to resist high temperatures to perform PCR, so Taq-polymerase was isolated (the DNA polymerase of the Termos Aquaticus bacteria, who lives at very high temperatures. We also need:
4 . DNA template thermostable DNA polymerase · primers · buffer · polymerase · dNTP later the Pfu Turbo was introduced, which also performed a 3' -+ 5' proofreading activityThe template sequence must be known in order to design its primers to initiate the synthesis of new DNA strand, DNA polymerase needs a primer designed to be complementary in order for a primer to bind properly it must
- have length of 18 - 25 nt
- GC% = 40% - 60%
- melting Temperature : T = 2 (A + T ) + 4 (G + C)
- no significant complementary within or between primers
PCR involves a number of cycles of different temperatures:
- DENATURATION at 94℃
- PRIMER ANNEALING at 55℃
- EXTENSION at 72℃
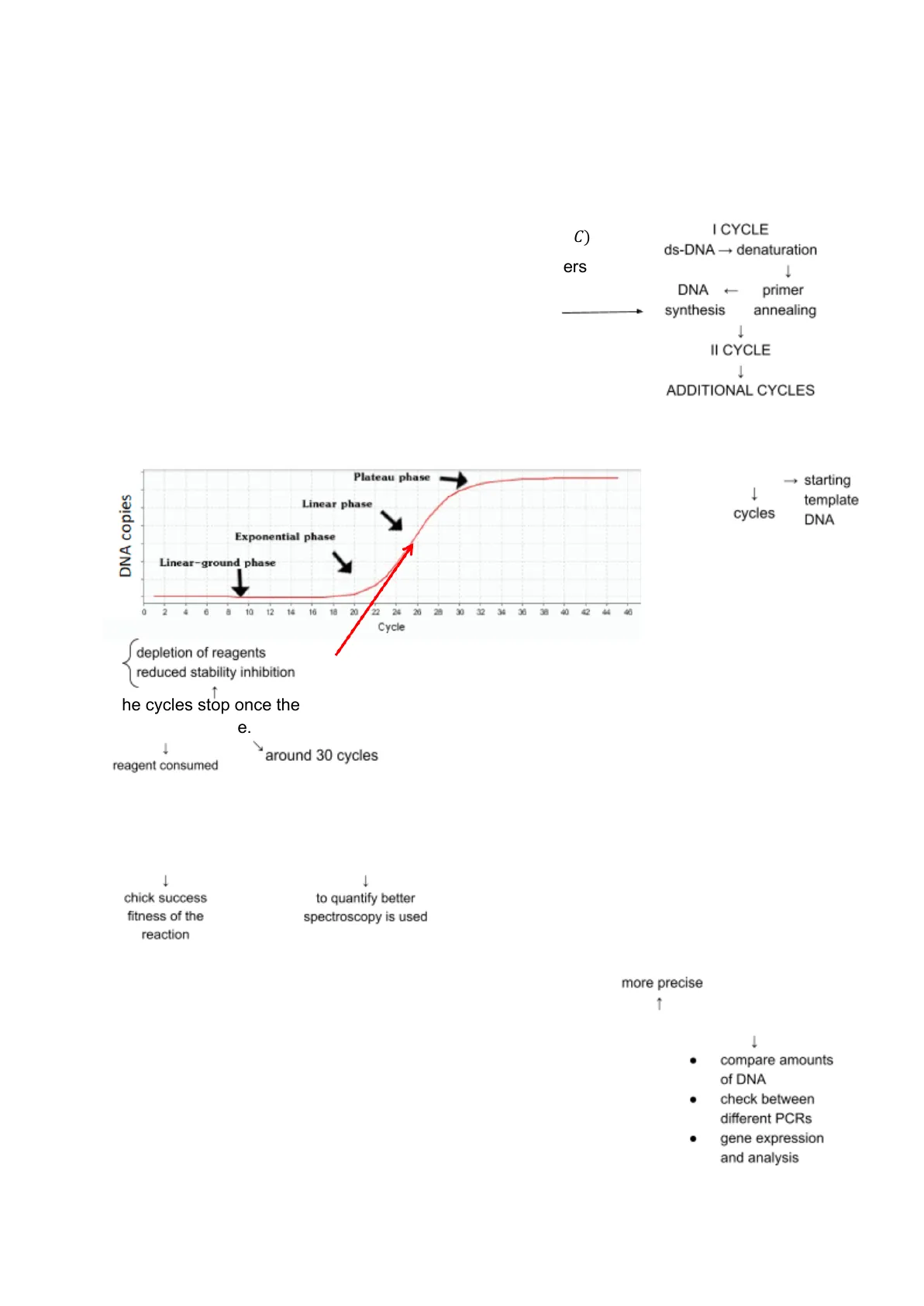
I CYCLE ds-DNA -+ denaturation - DNA + primer synthesis annealing 1 II CYCLE 1 ADDITIONAL CYCLES The PCR process can be done for many cycles at which the quantity of the desired strand increases exponentially. ↓ Plateau phase DNA copies Linear phase Exponential phase Linear-ground phase O 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 36 38 40 42 44 46 Cycle (2" - 2n) x - starting - template cycles DNA The cycles stop once the plateau phase is reached: after some time the DNA copies do not increase anymore.
reagent consumed 1 around 30 cycles The amount of PCR product is proportional to the amount of input DNA To know how much PCR product we had, ethidium bromide was used, which is an intercalating agent used as a fluorescent target. The fluorescence intensity is proportional to the sample quantity. It is then made run on electrophoresis gel.
1 chick success fitness of the 1 to quantify better spectroscopy is used reaction There are two types of PCR: more precise
- CONVENTIONAL (qualitative): only the result is quantitated 1
- REAL-TIME (quantitative): all cycles are quantitated while the reaction is processing To perform real-time PCR we can use another type of fluorescent DNA intercalator called SYBR green. The fluorescence emitted is then measured by an instrument that captures it and gives it a value proportional to DNA quantity in the eppendorf. - . compare amounts of DNA · check between different PCRs · gene expression and analysis depletion of reagents reduced stability inhibition
One of the most used applications of real-time PCR is gene expression analysis (how many RNAs are produced by a gene). RNA is not a substrate of TaqDNA polymerase. uses of rtPCR: To amplify RNA, it must be converted into DNA complements (cDNA). This conversion is carried · gene expression analysis · disease diagnosis · food testing · animal or plant breeding · forensics out by an enzyme able to use RNA as a template to synthesize DNA, called reverse transcriptase (RT). Then the produced DNA is amplified by PCR -> reverse transcription PCR
1 RT enzyme: · RNA template -> DNA synthesis RT PCR: · removes RNA from DNA strands 1) mRNA + primer 2) cDNA 3) DNA amplification depends on RNA type 1 The RT enzyme needs primers to work:
- oligo dT -> binds to the poly-A tail of mRNAs
- hexanucleotide hexamers -> random primers used when the sequence of RNA is not known internal primers
- specific oligo primer -> the sequence is known, a specific piece is wanted
DNA Cloning
To study a specific DNA sequence experimentally, it is necessary to generate enough copies for laboratory handling. PCR is a way to do so in vitro only, while DNA cloning requires a cell to replicate a specific gene info, for example a bacteria. To do so we require a vector to put our gene and transfer it to the host organism; the most simple example of a vector is the plasmid.
1 different vectors are used based on their insertion capacity 1 DNA cloning allows the take out of sequences of the genome and their re-insertion in another organism Plasmids are the most used vector as it is: · easy to handle · contains ORI · resistance to antibiotics( selection / discrimination mechanism) . recognized by many types of restrictions and enzymes Restriction enzymes are used to cut specific sequences at specific sites and cut vectors at the same specific site, in order for them to easily be inserted.
+ the plasmid is then inserted inside the bacteria and is used as molecular biofactory transformation amplification · synthesizes a second DNA strand on DNA template · found in retrovirusesThe sequences inserted in the vectors can also be transcriptionally controlled by their promoters; there are different kinds of promoters: · constitutive: gene expressed ubiquitously · tissue-specific · inducible/repressive cloning vector: DNA seq storage expression vector: DNA storage and use/analysis of expression of the gene To check the expression of genes, reporter genes are used, which encode for a product easy to detect (ex fluorescence). ↓ STEP 1. Select specific sequences (with specific restriction sites) are amplify them with PCR desired restriction site 4 PCR can be used to add the by adding it to the primer STEP 2. Insert the DNA by digesting with the same restriction enzyme and ligate our sequence with the complementary and of the plasmid STEP 3. Ligation of the sticky ends(H-bonds) STEP 4. Transformation of the plasmid inside the host bacterium changes or electrical discharges with chemicals and temperature STEP 5. Selection of the recombinant Clones using markers empty bacteria (ex. antibiotic resistance) empty plasmid recombinant plasmid PCR and cloning can be used together to produce mutations at specific DNA sites:
- deletion => obtained by putting reverse primers that exclude a central part of our gene
- single-point => obtained by using a primer containing a mutation Deletion ) - X Ligasi Single point mutation
Nucleic Acids Hybridization
-NUCLEIC ACIDS HYBRIDIZATION RNA and DNA can be denatured and renatured, meaning that with the increase of temperature, the hydrogen bonds of the nitrogen bases are disrupted leaving us with the single-stranded nucleotide chain; by going back to the original temperature the bonds are restored. Thanks to this it was easy to anneal the natural strand with an artificial one, generating heteroduplex.
(A) Denaturation by heating + Hybridization by cooling down temperature (B) denaturazione ibridazione (annealing) + + + + + + + OMODUPLEX ORIGINALI ETERODUPLEX ARTIFICIALI Hybridization technologies are used in many processes like:
- Northern and Southern blot, in which a labeled probe is made anneal to a set of sample present on a membrane. Only the probe corresponding and annealing strongly enough remain attached and are not washed away.
- Microarray, in which instead the probes are on the membrane and are made anneal with labeled samples. Those samples not annealing are then washed away.
Southern Blot Procedure
Southern blot is a method of detection of a specific DNA sequence among DNA samples. The procedure follows these steps:
- genomic DNA isolation
- digestion with restriction enzymes (short fragments)
- electrophoresis on gel of the DNA fragments -> separation of fragments
- denaturation of complementary DNA filaments
- transfer from gel to a membrane
- hybridization with a probe (labelled in radio) -> expose to X - ray corresponding to the complement of the gene interest
Gene-expression (and differential gene expression) is greatly studied as it plays a continuous and important role:
- TISSUES : different genes expressed => different cell type
- DEVELOPMENT : several genes must be expressed in different occasions
- SPATIAL REGULATION : embryo -> fetus
Northern Blot Procedure
Northern blot is a method used in molecular biology for detection of a specific transcript in RNA samples. Procedure:
- RNA extraction
- removal of secondary structures diffusion of
- electrophoresis charge
- transfer of the RNA into a membrane
- hybridization with labeled complementary probe
In both Northern blot and Southern blot; labeled probes are used to determine the presence and the amount of nucleic acids and they consist in a specific sequence complementary to the DNA or RNA target in the biological sample analyzed. Hybridization consists in a paring of single stranded molecules coming from two different sources:
- DNA or RNA target
- probe
1 hybridization is an extremely specific molecular recognition strategy Labeling is a basic technique that allows us to determine the position of a particular nucleic acid molecule. Labeled molecules emit a signal that can be caught. Labeled probes are used to determine the presence and the amount of nucleic acids; they consist of a specific sequence complementary to the DNA or RNA.
4 · TERMINAL LABELLING 1 DNA LABELLING · INTERNAL LABELLING . . RNA LABELLING · RADIOACTIVE TRACERS . NON-RADIOACTIVE TRACERS (fluorescence, chemiluminescence) 32P ex. y -ATP (terminal radioactive) The labeled probe and the target sequence are put together for hybridization for 16 hours. After that time they are washed away several times, in order for the unspecific probes to detach (help by high temperature and low salt, that are not favorable to unspecific binding). The population of all mRNAs transcribed by a cell is called TRANSCRIPTOME. It confers morphological and functional characteristics. Transcriptomics aims at the analysis of the expression of a large number of genes or of the entire transcriptome of the cell. The microarray technique is able to show the expression of several genes. Synthetic single stranded DNAs are spotted on a glass or silicon side (micro = chip). One oligonucleotide is produced for every 100-150 bp of DNA sequence in a sequential manner (array) resulting in a tiling array of DNA sequences.