B1.2 Proteins: Amino Acid Sequence, Structure, and Function
Slides from University about B1.2 Proteins. The Pdf explores proteins, their structure, and function, detailing the generalized structure of an amino acid and secondary protein structures like alpha-helix and beta-pleated sheet. This University Biology material is suitable for self-study.
See more40 Pages

Unlock the full PDF for free
Sign up to get full access to the document and start transforming it with AI.
Preview
Proteins: Amino Acid Sequence and Diversity
B1.2 Proteins SL/HL B1.2.1-B1.2.12 What is the relationship between amino acid sequence and the diversity in form and function of proteins? How are protein molecules affected by their chemical and physical environments?
Generalized Structure of an Amino Acid
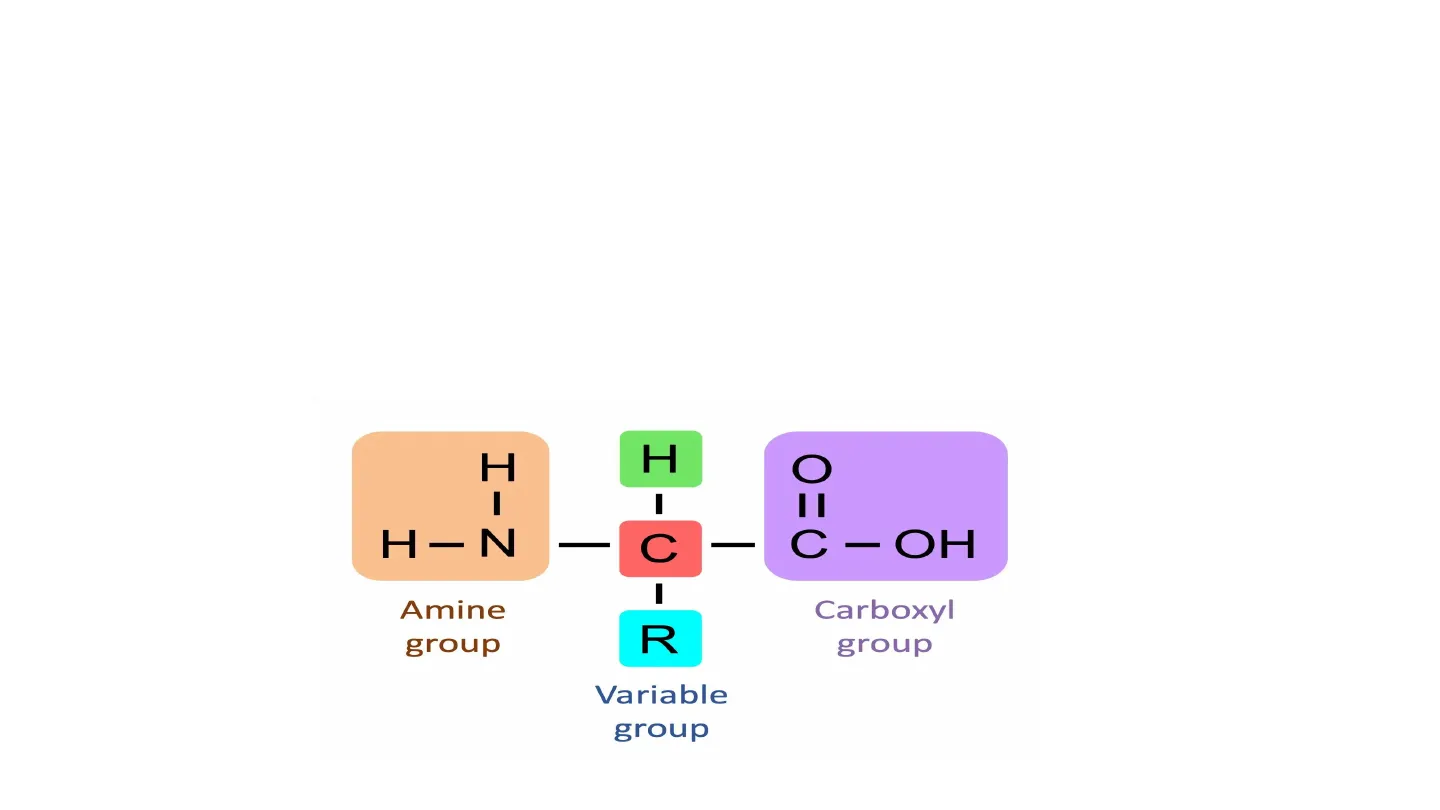
B.1.2.1 Generalized Structure of an amino acid 1 Proteins are composed of long chains of recurring monomers called amino acids . Each amino acid contains a central alpha carbon linked to an amine group, carboxyl group, a variable group and a hydrogen atom There are 20 different amino acids which are universal to all living organisms . Each type of amino acid differs in the composition of their variable side chain (denoted 'R' H H C I - I H -N C - C -OH - I Amine group R Carboxyl group Variable group
Condensation Reactions and Polypeptide Chains
Dipeptide Formation through Condensation
B1.2.2 Condensation reactions forming dipeptides and longer chains of amino acids 2 Amino acids can be covalently joined together via condensation reactions to form a dipeptide and water . The bond that is created is called a peptide bond and forms between the amine and carboxyl groups of adjacent amino acids Long chains of covalently bonded amino acids are called polypeptides and these chains can be broken down via hydrolysis reactions (requires water to reverse the process)
Dipeptide Formation Diagram
Dipeptide Formation 3 H H H 0 H H H 0 H 0 H H N C C + N C C N C C N C C + 0 H 0 H H 0 H 0 -H H R R R Peptide R Bond I 0-0-I C + Z U F O-H H I-0-₡ -O-H water removed H 0 Z I I-0-₡ V=0 I-Z I-0-₡ 0-0- H H peptide (amide) linkage +H2O F 1 Z - 0 H4
Dietary Amino Acid Requirements
Essential, Non-Essential, and Conditional Amino Acids
B1.2.3 Dietary requirements for amino acids Amino acids can be either essential, non-essential or conditional according to dietary requirements
- Essential amino acids cannot be produced by the body and must be present in the diet.Nine of the 20 aa are essential in humans. . Non-essential amino acids can be produced by the body (from other amino acids) and are therefore not required as part of the diet . Conditional amino acids can be produced by the body, but at rates lower than certain conditional requirements In other words, they are essential at certain times only (such as during pregnancy, infancy or illness) . A shortage of one or more essential amino acids in the diet will prevent the production of specific proteins . This is known as protein deficiency malnutrition and the health effects will vary depending on the amino acid shortage.
- Certain diets (e.g. vegan) require particular attention to ensure essential amino acids are consumed and malnutrition is avoided
Amino Acid Classification Table
Essential Conditionally Non-Essential Non-Essential Histidine Arginine Alanine Isoleucine Leucine Methionine Glycine Proline Phenylalanine Threonine Serine Tryptophan Tyrosine Valine Lysine Asparagine Asparatate Cysteine Glutamate fastbleep) Glutamine5
Peptide Chain Variety and Protein Diversity
Infinite Variety of Peptide Chains
B1.2.4 Infinite variety of possible peptide chains AS most natural polypeptide chains contain between 50 - 2000 amino acid residues, organisms are capable of producing a huge range of possible polypeptides
- Consequently, proteins are a very diverse class of compounds and may serve a number of different roles within a cell The number of possible amino acid sequences can be calculated starting with dipeptides. Both amino acids in a dipeptide can be any of the 20, so there are 20 x 20 possible sequences (20 2 ). There are 20 x 20 x 20 possible tripeptide sequences (20 3 ). For a polypeptide of n amino acids, there are 20 n possible sequences. The number of amino acids in a polypeptide can be anything from 20 to tens of thousands. For example, if a polypeptide has 400 amino acids, there are 20 400 possible amino acid sequences. This is an incredibly large number, and some online calculators simply express it as innity. Given that the number of amino acids in a polypeptide can be tens of thousands, the number of possible sequences is effectively infinite. But only an extremely small proportion are made by an organism. This is the organism's proteome.
Amino Acid R-Group Classification
NON-POLAR H O O O H3N+-C-C 1 o CH2 SH Cysteine (Cys / C) Proline (Pro / P) H O H O -O H3N+- C - C H3N+-C-C CH2 / CH2 H3C - CH CH2 S 1 CH3 Methionine (Met / M) O H3N+- C-C O H3N+- C - C CH2 HN Tryptophan ( Trp / W) Phenylalanine (Phe / F) + CHARGE H3N+- C- C O H3N+-C- C CH2 F O CH2 CH2 CH2 CH2 CH2 NH CH2 1 C = NH2+ NH3+ NH2 Lysine (Lys / K) Arginine (Arg / R) O I-U 1 O NH NH+ Histidine ( His / H) POLAR H H3N+-C- C O CH2 H3N+-C-C H3N+- C -C 1 O CH CH2 I OH OH CH3 OH Tyrosine ( Tyr / Y ) H O = H3N+-C- C O H3N+- C -C 1 O CH2 1 -0 NH2 Glutamine (Gin / Q) - CHARGE H H O H3N+- C- C CH2 1 CH2 CH- I C Aspartic Acid (Asp / D) == H3N+-C- 0 1 O CH2 1 CH2 C O O NH2 Asparagine (Asn / N) I-U- I-U-I -O H3N+- 1 0 H3N+-C-C / 0- O CH3 CH3 CH3 Glycine (Gly / G) Alanine (Ala / A) Valine (Val / V) H3N+- C - C. H O CH CH2 CH3 CH3 CH3 Leucine (Leu / L) Isoleucine ( lle / 1) H3N+-C -C H O CH 1 H O I-U Serine (Ser / S) Threonine (Thr / T) O 1 H2N+-C -C HỌC CH2 CH2 I-U- O O CH2 H O Glutamic Acid ( Glu / E)Functions of Proteins 6
Functions of Proteins
- Structure (collagen, spider silk)
- Hormones (insulin, glucagon)
- Immunity (antibodies)
- Transport (protein channels)
- Sensations (rhodopsin)
- Movement (actin, myosin)
- Enzymes amylase) (Rubisco, Rubisco, actin & myosin immunoglobulin (antibody) insulin hormonal contractile protection hemoglobin rhodopsin PROTEINS: Types and Functions receptor transport enzyme storage! 3+ Fe spider silk structural ferritin RUBISCO
Effect of pH and Temperature on Protein Structure
Protein Denaturation
B1.2.5 Effect of pH and temperature on protein structure 7 Denaturation is a structural change in a protein that results in the loss (usually permanent) of its biological properties
- Because the way a protein folds determines its function, any change or abrogation of the three dimensional structure will alter its activity Denaturation of proteins can usually be caused by two key conditions - heat (high temperatures) and pH DENATURATION Fleck Folded Protein Unfolded Protein
Temperature and pH Effects on Proteins
B1.2.5 Effect of pH and temperature on protein structure cont .. 8 Temperature:
- High levels of thermal energy may disrupt the hydrogen bonds that hold the protein together
- As these bonds are broken, the protein will begin to unfold and lose its capacity to function as intended
- Temperatures at which proteins denature may vary, but most human proteins function optimally at body temperature (~37ºC) pH:
- Amino acids are zwitterions, neutral molecules possessing both negatively (COO=) and positively (NH ) charged regions
- Changing the pH will alter the charge of the protein, which in turn will alter protein solubility and overall shape
- All proteins have an optimal pH which is dependent on the environment in which it functions (e.g. stomach proteins require an acidic environment to operate, whereas blood proteins function best at a neutral pH)
Effect of pH on Amino Acids
9 R H C +H3N COOH H+ R H H C +H3N COO- = H H C H2N COO- H+ Both groups protonated (Positive charge) Zwitterion (Neutral charge) Both groups deprotonated (Negative charge) Effect of pH on Proteins
Chemical Diversity of R-Groups
R-Group Diversity and Protein Form
RHL B1.2.6 Chemical diversity in the R-groups of amino acids as a basis for the immense diversity in protein form and function The 20 amino acids that ribosomes use to make polypeptides are very varied in the chemical nature of their R-groups. The elements present in the R-groups are shown in Table 2. When amino acids are linked up into a polypeptide, their amine and carboxyl groups are used to make peptide bonds. This leaves an amine group (-NH2 ) at one end of the chain and a carboxyl group (-COOH) at the other end. The hydrogen atom attached to the alpha carbon atom of each amino acid has little effect on the properties of the polypeptide; it is the R-groups that determine the chemical characteristics. Some of the R-groups are hydrophobic and some hydrophilic. Of the hydrophilic R-groups, some are polar and others become charged (+ or -) by acting as an acid or a base. This broad diversity of R-groups allows living organisms to make and use an amazingly wide range of proteins. Some of the differences between R-groups are shown in Table 3
Elements in R-Group Table
Elements in R-group Number of amino acids Honly 1 Cand Honly 5 C, Hand S only 2 C, Hand Nonly 5 C, Hand O only 5 C,H, Nand O 2 Table 2 Variation in R-groups of amino acids
Amino Acid Classification by R-Group
Classification of amino acids Eleven R-groups are hydrophilic Nine R-groups are hydrophobic with between zero and nine carbon atoms Seven R-groups can become charged Three R-groups Six R-groups contain rings do not contain rings Four hydrophilic R-groups are polar but never charged Four R-groups act as an acid by giving up a proton and becoming negatively charged Three R-groups act as a base by accepting a proton and becoming positively charged Table 3 Classification of amino acids
Impact of Primary Structure on Protein Conformation
Protein Structure Levels: Primary
B1.2.7 Impact of primary structure on the conformation of proteins The structure of proteins has four levels of complexity: primary, secondary, tertiary and quaternary. Primary structure is the linear sequence of amino acids in a polypeptide. The backbone of a polypeptide is a repeating sequence of atoms linked by covalent bonds ( C C N C C N and so on). The bond angles are all tetrahedral and there can be rotation about the bonds between the alpha carbon atoms and adjacent nitrogen and carbon atoms. This allows polypeptides to fold into almost any three-dimensional shape.primary structure https://www.youtube.com/watch?v=piXHivrTT-E The three-dimensional arrangement of atoms in a polypeptide or protein is its conformation. Most polypeptides self-assemble into a specific conformation determined by the sequence of amino acids and their R-groups. The conformation of proteins determines their functions and through this the behaviour of cells. This is why protein conformation is of great interest to biologists
Primary Structure Diagram
Primary Structure https://www.youtube.com/watch?v=hok2hyED9go 5 R C-a N N-terminus C C-a N C C-a C-terminus H peptide bond (no rotation) rotation about carbon bonds H
Secondary Structure of Proteins
Pleating and Coiling in Secondary Structure
B1.2.8 Pleating and coiling of secondary structure of proteins At regular intervals along a polypeptide chain there are C=O and N -H groups. They are what remains of carboxyl and amine groups after they have been used to make peptide bonds. Both C=O and N -H are polar, with the oxygen having a slight negative charge and the hydrogen having a slight positive charge. Due to this polarity, hydrogen bonds can form between these groups. Although hydrogen bonds are individually weak, the frequency of C=O and N -H groups along polypeptide chains allows many of them to form and collectively they are strong enough to stabilize distinctive conformational structures within protein molecules.