Regulation and Inhibition of Enzymes: Understanding Enzyme Control
Slides from Biocd 1556 about Regulation and Inhibition of Enzymes. The Pdf, a detailed presentation for university-level Biology students, explores enzyme control mechanisms, reaction velocity factors, and Michaelis-Menten kinetics, providing a comprehensive overview of the subject.
See more12 Pages
Unlock the full PDF for free
Sign up to get full access to the document and start transforming it with AI.
Preview
Regulation and Inhibition of Enzymes
BIOCD 1556 Lecture 3 Dr. Susan Viselli Regulation and Inhibition of Enzymes Introduction Enzymes are protein catalysts that increase the rate of biological reactions without being changed in the overall process. The goal in an enzyme-catalyzed reaction is for an enzyme to convert its substrate into products. Most enzymes function inside cells, oftentimes within cellular organelles. Biochemical processes happen at the cellular level and are regulated by the function of enzymes. Regulation of the reaction velocity of enzymes is therefore essential for the coordination of metabolic processes. Mechanisms for regulation of enzymes in metabolic pathways include: substrate availability, product inhibition, allosteric control, covalent modification, and synthesis or degradation of the enzyme. Any substance that can diminish the velocity of an enzyme-catalyzed reaction is called an inhibitor. Enzymes are normally inhibited by metabolic and dietary constituents and may be also be abnormally inhibited by toxins and pharmacologically inhibited by drugs. Many commonly dispensed drugs in the United States function by inhibiting enzymes.
Terminal Objective
TERMINAL OBJECTIVE: To understand the mechanisms of enzyme regulation and the mechanisms of enzyme inhibition.
Enabling Objectives
- Describe factors that may affect the rate of enzyme-catalyzed reactions.
- Describe the importance of Km and be able to determine the Km and Vmax from both Michaelis-Menten and Lineweaver-Burke plots.
- Compare competitive and noncompetitive enzyme inhibition and understand the effects of competitive and noncompetitive inhibitors on Km and Vmax.
- Explain mechanisms for regulating enzyme activity including substrate availability, product inhibition, allosteric control, covalent modification, and synthesis or degradation of enzyme.
Reading: Lippincott's Illustrated Reviews: Biochemistry, pages 53-64.
Substrate Product Active Site ENZYME E + S ES 1 E + PI.
Overview of Enzyme Function
Energy Changes During Reaction
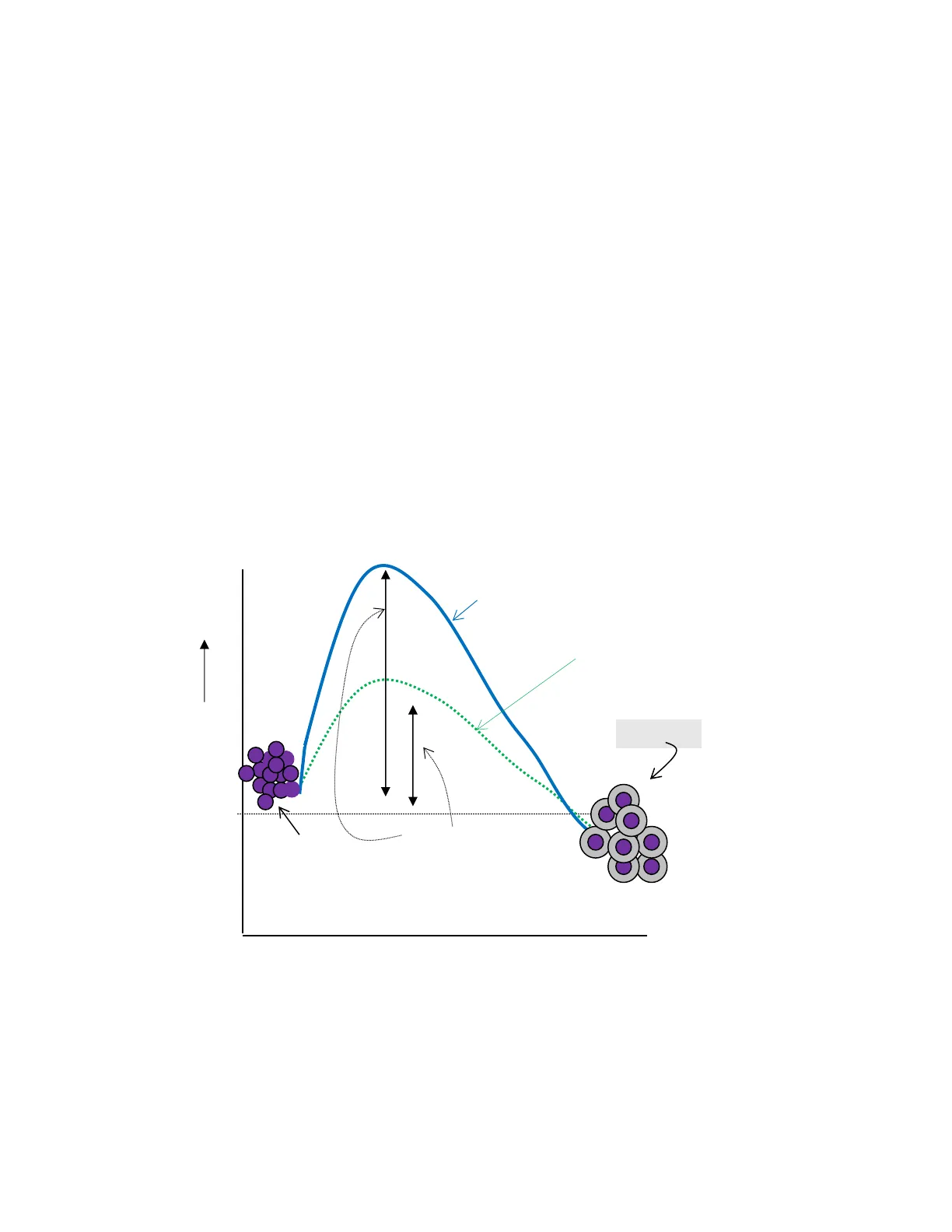
A. Energy changes occurring during the reaction An energy barrier separates reactants and products. The free energy of activation is the energy difference between the reactants and the transition state, a high-energy intermediate that occurs during product formation. For molecules to react they must contain sufficient energy to overcome the energy barrier of the transition state. The rate of the reaction is determined by the number of molecules that have sufficient energy to overcome the barrier. In general, the lower the free energy of activation, the more molecules will have sufficient energy to reach the transition state, and the faster the rate of the reaction. In the absence of an enzyme, only a small proportion of the reactant molecules may possess enough energy to achieve the level of the transition state. An enzyme allows a reaction to proceed rapidly under conditions prevailing in the cell by providing an alternate reaction pathway with a lower free energy of activation.
Transition State Un-catalyzed Reaction Catalyzed Reaction Free Energy Products Reactants Free Energy of Activation Progress of Reaction >
Chemistry of the Active Site
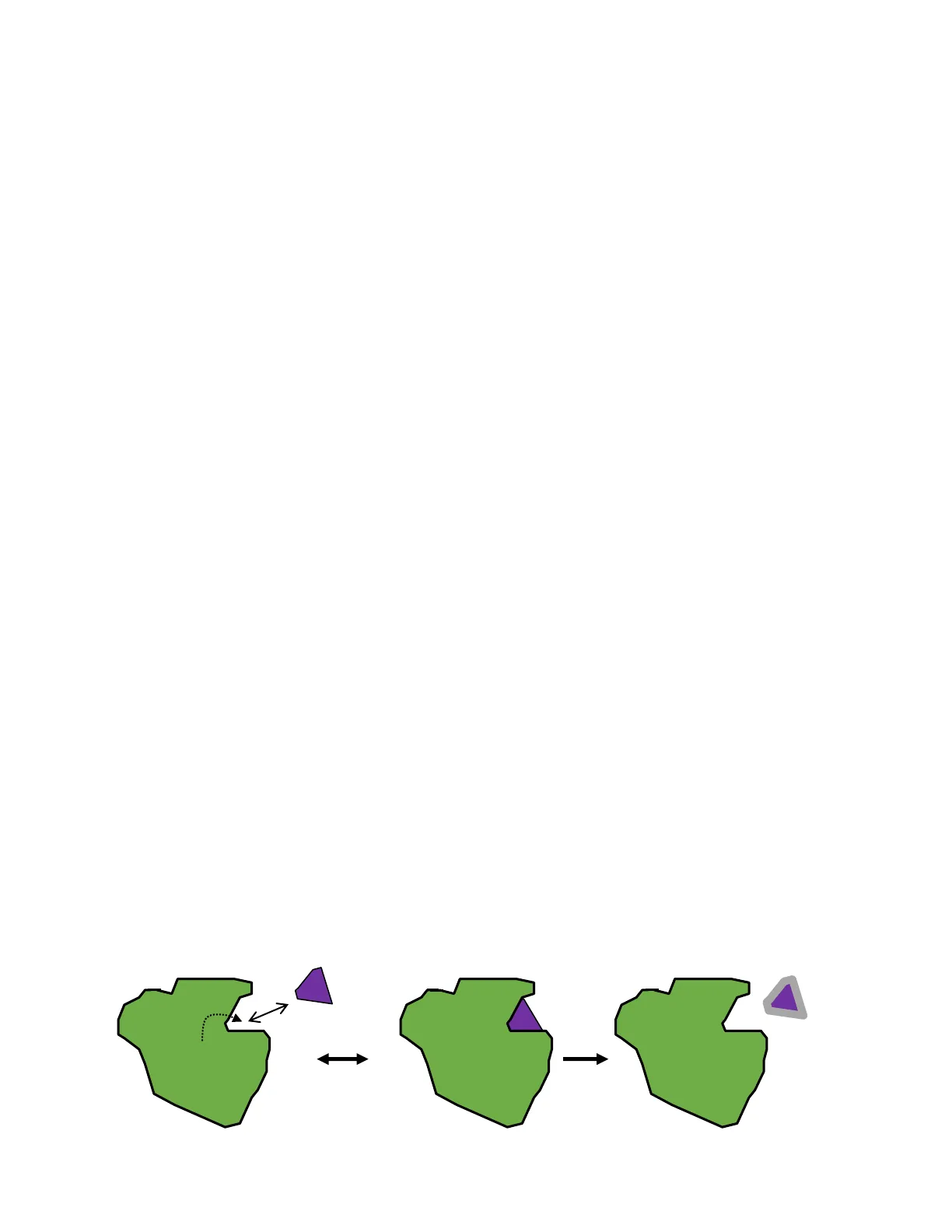
2B. Chemistry of the active site The active site is a complex molecular machine that facilitates conversion of substrate to product.
Substrate 5 Active site Enzyme Copyright ( 2008 Wolters Kluwer Heath | Lippincott Williams & Wilkins The active site of an enzyme acts as a flexible molecular template that binds the substrate and stabilizes the substrate in the transition state on its way to becoming product. The active site can provide catalytic groups to enhance the probability that the transition state is formed. This may involve general acid-base catalysis where amino acid residues accept or donate protons. In other cases there is the transient formation of a covalent enzyme-substrate (ES) complex.
Function of Enzymes
C. Function of Enzymes An enzyme holds its substrate in a state intermediate between its original reactant state and that of product. Enzyme helps the substrate achieve the transition state. (See depiction in the figure to the right.) Note that when enzyme and substrate initially combine there is a dip in free energy from where S alone was shown. This brief interaction stabilizes the substrate but the lower energy level is not maintained. ES is described as having a "brief" covalent interaction. Enzyme then helps the substrate achieve its transition state. After reaching the transition state the now altered substrate (product) has lower levels of free energy. Product is an altered form of the substrate.
energy T S ES Substrate Free P Progress of reaction T Enzyme S ES Free P Progress of reaction Enzyme- substrate complex (ES) Enzyme Free energy ES P Transition state Progress of reaction Free energy T S ES Product (P) P Progress of reaction Copyright @ 2008 Wolters Kluwer Health | Lippincott Williams & Wilkins 3 T* S energyD.
Factors Affecting Reaction Velocity
Factors Affecting Reaction Velocity The rate (or velocity) of a reaction is determined by the number of substrate molecules that are converted to product per unit time. (umoles / second is often used for rates)
Substrate Concentration and Vmax
1. Substrate Concentration For a range of substrate concentrations, reaction velocity will increase with increasing substrate concentration, until a maximal velocity (Vmax) is reached. The leveling off of the reaction rate at high substrate concentrations reflects the saturation with substrate of all available binding sites on the enzyme. Therefore, at high substrate concentration, all the binding sites on the enzyme are saturated with substrate.
Vmax Reaction velocity [Substrate concentration] After Vmax is reached, adding more substrate will not increase the rate of the reaction.
Temperature Effects on Enzymes
2. Temperature - up to a point, increasing temperature will increase the rate of reaction. When temperature becomes too high, it can adversely affect the structure and function of the enzyme.
pH Effects on Enzymes
3. pH - Enzymes function best within a certain range of pH values, based on properties of the amino acids that comprise the enzyme's structure. Many enzymes have optimum pH close to pH of 7 (neutral), while others function best under acidic (low pH) or basic (high pH) conditions.
Reaction Model: Michaelis-Menten Equation
II. Reaction model The Michaelis-Menten equation describes the relationship between reaction velocity and substrate concentration using a simple model that accounts for most features of enzyme-catalyzed reactions. In this model, the enzyme combines reversibly with its substrate to form a complex that will break down into product and regenerate the free enzyme. S is the substrate E is the enzyme ES is enzyme-substrate complex E + S k1 k.1 ES +E + P k2 k1, k-1, and k2 are rate constants 4A.
Michaelis-Menten Equation Details
Equation The Michaelis-Menten equation is Vo = Vmax [S] Km + [S] Vo = initial reaction velocity Vmax = maximal velocity Km = Michaelis constant (k_1 + k2)/k1 [S] = Substrate concentration
Vmax Reaction velocity (v0) Vmax 2 Km [Substrate]
Conclusions on Michaelis-Menten Kinetics
B. Important conclusions about Michaelis-Menten kinetics The Michaelis constant, Km, reflects the affinity of the enzyme for a particular substrate. Km is expressed in terms of substrate concentration and is numerically equal to the substrate concentration at which the reaction velocity equals 1/2 Vmax. Small Km reflects a high affinity of enzyme for substrate; low [S] needed to reach 1/2 Vmax Large Km reflects a low affinity of enzyme for substrate; high [S] needed to reach 1/2 Vmax In this example (on right) Enzyme 2 has a greater Km than Enzyme 1 and therefore Enzyme 1 has more affinity for the substrate.
Vmax Reaction velocity (Vo) Enzyme 1 Enzyme 2 Vmax 2 0 0 A [Substrate] Km, Km2 Large Km of enzyme 2 reflects a low affinity of enzyme for the substrate. Small Km for enzyme 1 reflects a high affinity of enzyme for the substrate. Copyright 2008 Wolters Kluwer Health | Lippincott Williams & Wilkins 5C.
Lineweaver-Burke Plot Analysis
Lineweaver-Burke Plot It can be somewhat difficult to determine Vmax and Km from a hyperbolic shaped curve because of the gradual upward slope of the hyperbolic curve at high substrate concentrations. Transformation of the Michaelis-Menten equation into a straight line can be done by plotting 1/Vo versus 1/[S] (a double reciprocal plot). This allows for an easier graphical determination of Km and Vmax. A Lineweaver Burke plot is a transformation of the Michaelis-Menten equation into a straight line by performing a "double reciprocal" plot, 1/ [S] vs. 1/ V .. 1 1 = Km + 1 Vmax VO 1 Vo Vmax[S] Vmax intercept on the x-axis is -1 / Km intercept on the y-axis is 1/ Vmax
-1 Km 1 [S]
Inhibition of Enzyme Activity
III. Inhibition of Enzyme Activity Inhibitor - any substance that can diminish the velocity of an enzyme-catalyzed reaction
Competitive Inhibition
A. Competitive Inhibition - the inhibitor binds to the same site on the enzyme that substrate normally binds; therefore, the inhibitor binds to the enzyme active site. Inhibitor and substrate "compete" for binding to the active site on enzyme. Malonate is an example of a competitive inhibitor. Succinate dehydrogenase catalyzes the oxidation of succinate to fumarate. Malonate is structurally similar to succinate (substrate) and competes for binding to the active site of the enzyme. The enzyme-malonate complex is unreactive. By increasing the concentration of succinate, the probability increases that the active site will be occupied by a substrate molecule instead of by an inhibitor.
Substrate (succinate) Competitive Inhibitor (malonate) Succinate Dehydrogenase 6Graphing the impact of a competitive inhibitor:
Vmax Reaction Velocity 1 v T 1 Vmax -1 -1 1 [S] Km Km With competitive inhibitor No inhibitor present Effect of competitive inhibitor on Vmax - not changed, at high [S], Vmax can be reached why? -- If enough substrate is added, the effect of the inhibitor can be overcome Effect of competitive inhibitor on Km - increased why? -- In the presence of a competitive inhibitor more substrate must be added to achieve 1/2Vmax.
HMG CoA reductase Active site OH ĆHa CH 11 HO O HO O HỌC OH OH HMG CoA (substrate) CH Lovastatin (competitive inhibitor) Copyright @ 2008 Wolters Kluwer Health| Lippincott Williams & Wilkins Statin drugs are examples of competitive inhibitors (see figure to the left) These cholesterol-lowering drugs (also called anti-hyperlipidemic agents) competitively inhibit HMG CoA reductase which catalyzes the first committed step in cholesterol synthesis. The outcome is that less cholesterol is produced within liver cells in the patient using the statin drug. 7 O CH Km Km [Substrate]