Human Physiology: Membrane Transport Mechanisms
Slides from Dipartimento Di Medicina Sperimentale E Clinica, Univpm about Human Physiology. The Pdf, a university-level Biology document from 2023, details membrane transport mechanisms, including passive and active transport, with clear diagrams and concise text.
See more48 Pages

Unlock the full PDF for free
Sign up to get full access to the document and start transforming it with AI.
Preview
Membrane Transport Overview
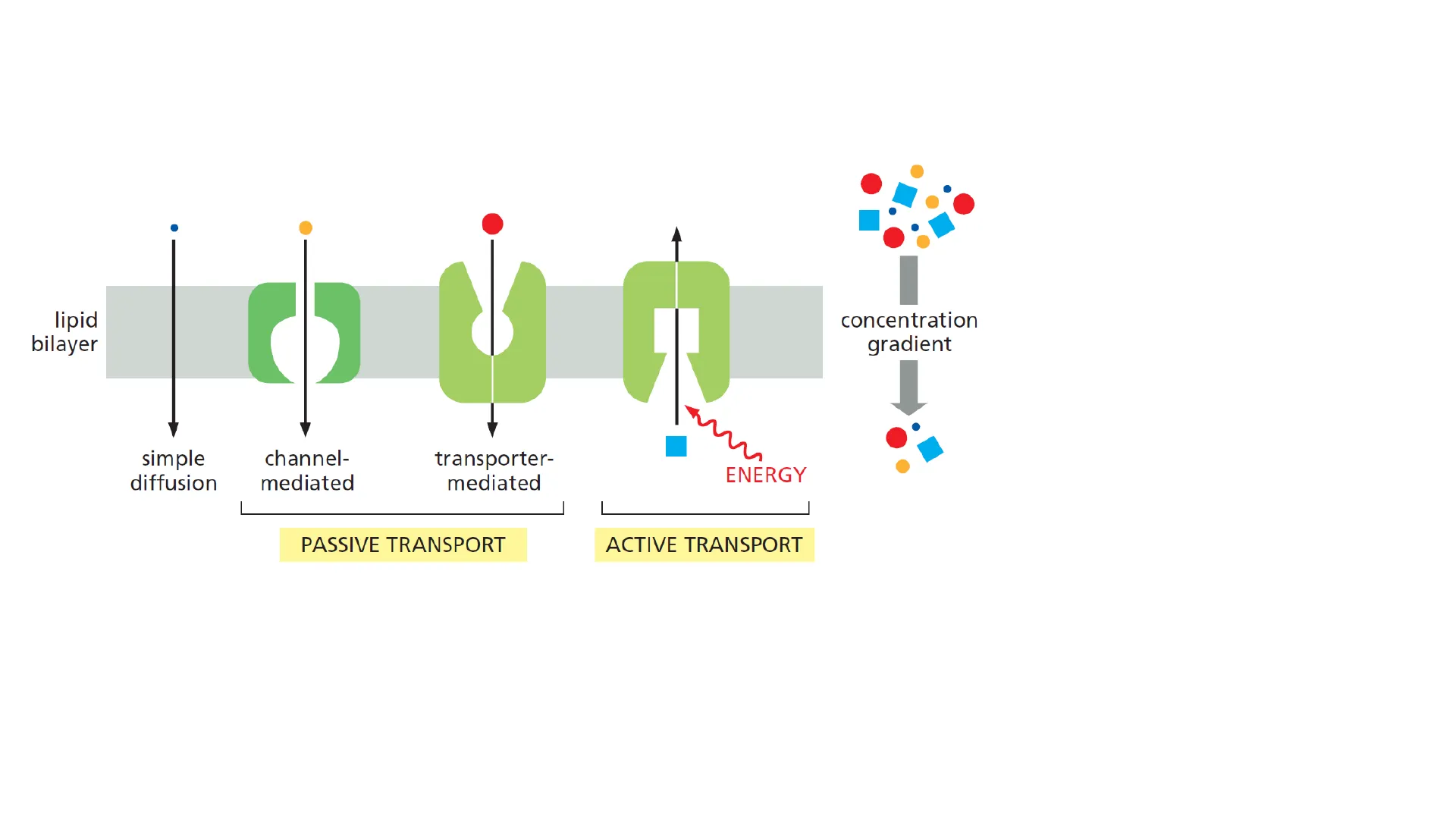
Human Physiology Simone Pifferi Dipartimento di Medicina Sperimentale e Clinica s.pifferi@staff.univpm.itMembrane transport lipid bilayer concentration gradient simple diffusion channel- mediated transporter- mediated ENERGY PASSIVE TRANSPORT ACTIVE TRANSPORT
- Diffusion
- Protein-mediated
- Ion channels
- TransportersMembrane transport
- Passive Down gradient (concentration/electric potential)
- Active Against gradient
Simple Diffusion Principles
Simple Diffusion Ji = Pi * (C1 - C2) Pi Permability Coefficient Transmembrane Flux high Permeability low Permeability P. P2<P P2 C1-C2 high permeability 02- . 102 1 - 10-2 H2O - -- 10-4 urea glycerol 10-6 tryptophan glucose 10-8 10-10 CI- K+ - 10-12 Na+- - 10-14 low permeability permeability coefficient (cm/sec)Simple Diffusion Ji = Pi * (C1 - C2) Dm Pi = Bi ΔΧ Bi Partition coefficient Dm Diffusion coefficient Ax Thickness of membrane HYDROPHOBIC MOLECULES O2 CO2 N2 steroid hormones SMALL UNCHARGED POLAR MOLECULES H2O urea glycerol NH3 LARGE UNCHARGED POLAR MOLECULES glucose sucrose IONS H+ Na+ HCO3 K+ Ca2+ CI- Mg2+
Protein-Mediated Transport
Ion Channels Structure
Protein-mediated transport · Ion channels Open Extracellular side 1 Cytoplasmic sideProtein-mediated transport Ion channels
Ion Channels Characteristics
•Protein-mediated transport · Ion channels · Selectivity · GateIon channels · Selectivity Ion P lon! /P Na Na+ 1 K+ 0.086 Ca2+ <0.01Ion channels Resting membrane potential Voltage amplifier and oscilloscope Voltage amplifier and oscilloscope Extracellular electrode Insert microelectrode V Extracellular electrode Nerve cell Nerve cell +60 L Insert electrode +30 I Vm (mV) 0 Resting potential 1 -30 -60 -90 Time
Ion Channels Gating Mechanisms
Ion channels • Gate Change in membrane potential + + + + - + + + + - + + + + Voltage-gatedIon channels • Gate Closed Open Ligand binding Extracellular side Cytoplasmic side Ligand-gated (extracellular or intracellular)Ion channels • Gate Direct activation through lipid tension Direct activation through structural proteins MechanosensitiveIon channels • Gate · Ankyrin domain Extracellular side TRPA1 TRPM8 TRPV4 TRPV3 TRPV1 TRPV2 C C C C C C - N -N N N -N * Receptor temperature range (℃) 0 10 20 30 40 50 60 Temperature-gated Cytoplasmic side NIon channels • Gate Na+ Light-gated
Aquaporins and Water Transport
Membrane transport · Aquaporins 13 types high permeability . 102 1 -- 10-2 H2O -- 10-4 urea glycerol 10-6 tryptophan glucose permeability coefficient (cm/sec) - 10-8 10-10 CI- K+ - Na+ 10-12 -10-14 low permeabilityAquaporins CNS Astrocyte end-foot AQP1 CSF AQP4 AQP5 AQP1 AQPO Aqueous Lens humor AOP3 Retina Optic nerve C 82898 Lacrimal gland- AQP5 AQP4 Olfactory epithelium Support cell Olfactory receptor neuron Pain C-fibres AOP1 DRG neurons (non- myelinated C-fibres) Lung Airways Alveolus -AQP4 -AQP5 AQP3 AQP4 Type I cell Type Il cell Erythrocyte AQP1 AQP9 Parietal cell AQP4 Lumen Lumen AQP3 Leukocytes Intestine AQP8 AQP3 AQP4 Adipocyte AQP7-0 Kidney Principal cell AQP2 Proximal tubule -AQP3 0 AQP75 53 AOP1 Loop of Henle Collecting duct Vasa recta Tumour Tumour cell Endothelium AQP1 Various AQPs Pillar cell Deiters cell Epidermis Stratum corneum Granular layer Acinus Duct AQP4 Endothelium Ependyma Pia Glia limitans externa Inner ear AQP4 Scala media Hensen cell Claudius cell Inner sulcus Inner hair cell Scala tympani AQP4 Eye Ciliary epithelium Lumen Choroid plexus CSF Salivary gland Spinous layer AQP3 Basal layer AQP4 AQP1 Skeletal muscle AQP4 Stomach Endothelium AOP1 AQP3 ConjunctivaAquaporins Loop a Loop c Loop e M7 M1 M2 M4 M5 M6 M8 M3 Loop b Loop d N terminus C terminus 1 2 4 3 8 AQP monomer 2.7 nmAquaporins Stimulation of baroreceptors or osmoreceptors Hypothalamus Pituitary gland Vasopressin release PCT TAL CD Urine HO HO Apical AQP1 AQP7 PCT Basolateral HO Blood Proximal convoluted tubule H.O HO AQP2 Endocytosis CD Translocation AQP3-1 -AQP4 HO HO Collecting duct
Protein-Mediated Transporters
Transporter Characteristics
Protein-mediated transport · Transported · Specificity · Saturation · CompetitionProtein-mediated transport · Transporters · Specificity · Saturation J = Jmax [S] KM + [S] 1.0 JIJmax 0.5 0.0 KM [S]Protein-mediated transport · Transporters · Specificity · Saturation J = Jmax [S] KM + [S] 1.0 J/Jmax 0.5- 0.0 KM KM [S]Protein-mediated transport · Transporters · Competition 1.0 +C J/Jmax 0.5 0.0 KM KM [S]
Transporter Mechanisms
Transporters transported molecule co-transported ion lipid bilayer UNIPORT SYMPORT ANTIPORT coupled transportTransporters Mechanism of transport solute OUTSIDE lipid bilayer INSIDE OUTWARD- OPEN OCCLUDED INWARD- OPENTransporters Passive solute . OUTSIDE INSIDE OUTWARD- OPEN INWARD- OPEN KM out = KM in [A] out [A]in Time Active solute OUTSIDE INSIDE OUTWARD- OPEN INWARD- OPEN > KMin KM out KM out < KM in [A] out [A]in Time
Facilitated Diffusion Transporters
Transporters Facilitated diffusion Expression KM M Insulin-dependence GLUT1 Red blood cells, many tissues 5 No GLUT2 Liver, ß-cells, intestine, kidney 7 No GLUT3 Neurons, glia 1.8 No GLUT4 Skeletal muscle, adipocytes 4.5 Yes GLUT5 Intestine 6 No GLUT6 Endoplasmic reticulum NoTransporters 100- max % of glucose transport velocity 80 GLUT3 60 50 40 GLUT2 20 [Glucose]. out (mM) KJ 4 6 KM3 10 12 14 16 M2 Physiological range of glycaemia
Active Transport Systems
Primary Active Transport
Transporters Active (against gradient) · Primary Directly use ATP · Secondary Use the electrochemical gradient of other solutes (created and/or maintained using ATP)Transporters ions small molecule H+ or (K+) or (Nat) or Ca++ lipid bilayer CYTOSOL P H+ H+ ADP ATP ADP + P ATP ADP ATP + ATP ADP + Pi Pi+ ADP ATP Pi P-type pump ABC transporter V-type proton pump F-type ATP synthase H+ H+
ABC Transporters
ABC Transporters ABC (ATP-binding cassette) MDR (multi drug resistance) C N ATP ATPABC Transporters CFTR (Cystic fibrosis transmembrane conductance regulator) TMD1 TMD2 N 1 R 2 C CFTR has the structure of ABC transporters but is an ion channel C1 O 1 O C2 1 O 2ABC Transporters CFTR (Cystic fibrosis transmembrane conductance regulator) Normal CFTR TMucus clearance Mucus Layer PCL Na+ Na* H2O Na H2O Apical surface CI- K Na+ Basolateral surface K+ K Na+ CI- CFTR mutation Mucus clearance Mucus Layer Na+ PCL Na+ H20 Na+ H2O Apical surface CI. K Na+ Basolateral surface "K+ -K+ Na+ CI-
Proton Pumps
Primary active transport H-ATPasi (proton pump) • V · F (ATP-synthase) F-ATPasi B a 8 ADP+P F1 ATP bb H+ Y Matrix 3 Fo a C H+ i Intermembrane space V-ATPasi B A A C ADP+P V1 B B E A -ATP G G H d D H+ F Cytosol a C Vo c C c Lumen H+
P-ATPase Family
Primary active transport P-ATPase Na+ Ca2+ Na-K pump PMCA K+ H-ATPase (V) K+ Ca2+ SERCA H ER Lys ABC-transporterPrimary active transport P-ATPase NH2 N a A P COOH Cytosol 10 9 8 7 65 4 321 S T N nucleotide P phosphorylation domain A actuator (phosphatase) T Transport unit S Support unit
Na+/K+ Pump
Primary active transport Na+/K+ pump 3 Na+ plasma membrane Na+ electrochemical gradient K+ electrochemical gradient CYTOSOL 2 (K+ P ADP ATP 800.000/3 millions copies per cells 40% of energy used by an organism is necessary to maintain the activity of Na+/K+ pump Electrogenic (hyperpolarize the membrane potential)Primary active transport Na+/K+ pump 1 ECF [Na+] high ATP ADP + energy 5 High-affinity binding sites for Na+ appear. 3 Na+ from ICF bind to high-affinity sites. ICF [Na+] low K-binding sites lose their affinity for K+ and release 2 K+ into ICF. ATPase is phosphorylated with Pi from ATP. P Protein changes conformation. Pi released. 4 2 K+ from ECF bind to high-affinity sites. 3 Na-binding sites lose their affinity for Na+ and release 3 Na+ into ECF. [K+] low P High-affinity binding sites for K+ appear. P [K+] high 2 P Protein changes conformation.Primary active transport Na+/K+ pump O O OH OH H H HO HO O O H OH HO" O H H Digoxin Ca++ Na+ Na+/Ca ** exchanger Na*/k+. K+ ATPase 1Ca++ T Na+ K+ 1 TN-C Ca" binding ÎInotropy
H+/K+ Pump
Primary active transport H+/K+ pump Lumen Interstitial space H+ H2O Na+ 3 Na1 2 K+ HC Parietal cell ATP K+ K+ HCO3 OH- H+ ATP CI- CA CO2 CO2 H2O
PMCA and SERCA Pumps
Primary active transport PMCA/SERCA [Ca2+]ex=1.3/1.2 mM [Ca2+] ] = 100 nM [Ca2+]reticulum=0.5/0.6 mM reticulum Extracellular space Ca2+ Ca2+ PMCA Ca2+ B ATP Ca2+ Ca2+ Cytosol [Ca2+], = 100 nM ADP ADP ADP ATP 2Ca2+ 2Ca2+ ATP B SERCA ER 2Ca2+ 2Ca2+
Secondary Active Transport
Secondary active transport It used the energy give by . Ionic gradients generated by primary active transport (Na+) · Gradients of organic molecules generated by metabolismSecondary active transport IV HCO, CI- K+ CI- 2Na+ 3Na+ Glucosio 2Ca2+ III Na Na+ Amino- acidi · H+ Na+ CI- K+ Na+ L Na+ 2CI- II I ) Organic molecules cotransporters II ) Symport Na+/other ions III ) Antiport Na+-dependent IV) Others
Neurotransmitter Transporters
Secondary active transport Symport 2Na+/Neurotransmitters GAT (GABA) GLYT (Glycine) DAT (Dopamine) SERT (Serotonin) EAAT (excitatory Amino acid transporters)Neurotransmitter transporters MONOAMINERGIC NEURON GABAERGIC NEURON VIAAT Monoamine transporter GABA transporter GAT1 GAT1 GLIAL CELL BGT1 GAT3 Monoamine receptors GABA receptors GLYCINERGIC NEURON VIAAT Glycine transporter GLYT2 GLYT2 GLIAL CELL GLYT1 Glycine receptors AVMAT SERT DAT NET O GLIAL CELLNeurotransmitter transporters GAT (GABA) GLYT (Glycine) DAT (Dopamine) SERT (Serotonin) solute OUTSIDE 1a 6a 5 4 2 3 8 7 9 10 11 12 lipio bilayer 14 1b 6b INSIDE -N C OUTWARD- OPEN OCCLUDED INWARD- OPENNeurotransmitter transporters EAAT extracellular HP2 12 3 4 5 6 7 8 HP1 NH2 COOH intracellular Transport Scaffold Outside Na+ Citrate Inside VCINDY Inward facing VCINDY Outward facingNeurotransmitter transporters Elevator mechanism Gate Transport domain Scaffold domain Gate