Appunti sul ciclo cellulare, struttura del cromosoma e mitosi
Documento sul ciclo cellulare. Il Pdf, un appunto di Biologia per l'Università, descrive il ciclo cellulare, la struttura del cromosoma e il processo di mitosi, incluse le sue fasi e la citocinesi, con un focus sul complesso APC.
Ver más14 páginas
Visualiza gratis el PDF completo
Regístrate para acceder al documento completo y transformarlo con la IA.
Vista previa
Ciclo Cellulare
The cell cycle begins when two new cells are formed by the division of a single parental cell and ends when one of these cells divides again into two cells.
It can be divided into two major phases based on cellular activities:
- Interphase: it's the period between cell divisions, the cell grows and engages in diverse metabolic activities.
0 G1, the cell grows and carries out normal metabolism, the organelles duplicate o S: DNA replication 0 G2: the cell grows and prepares for mitosis
G1 and G2 are called "gap" phases: G1 separates S phase from the preceding M phase, G2 separates the end of the S phase from the next M phase
- M phase: process of cell division o Mitosis o Cytokinesis
The M phase usually lasts less than an hour while the cell cycle takes about 18-24 hours. Interphase may extend for days, weeks or longer, depending on the cell type and the conditions.
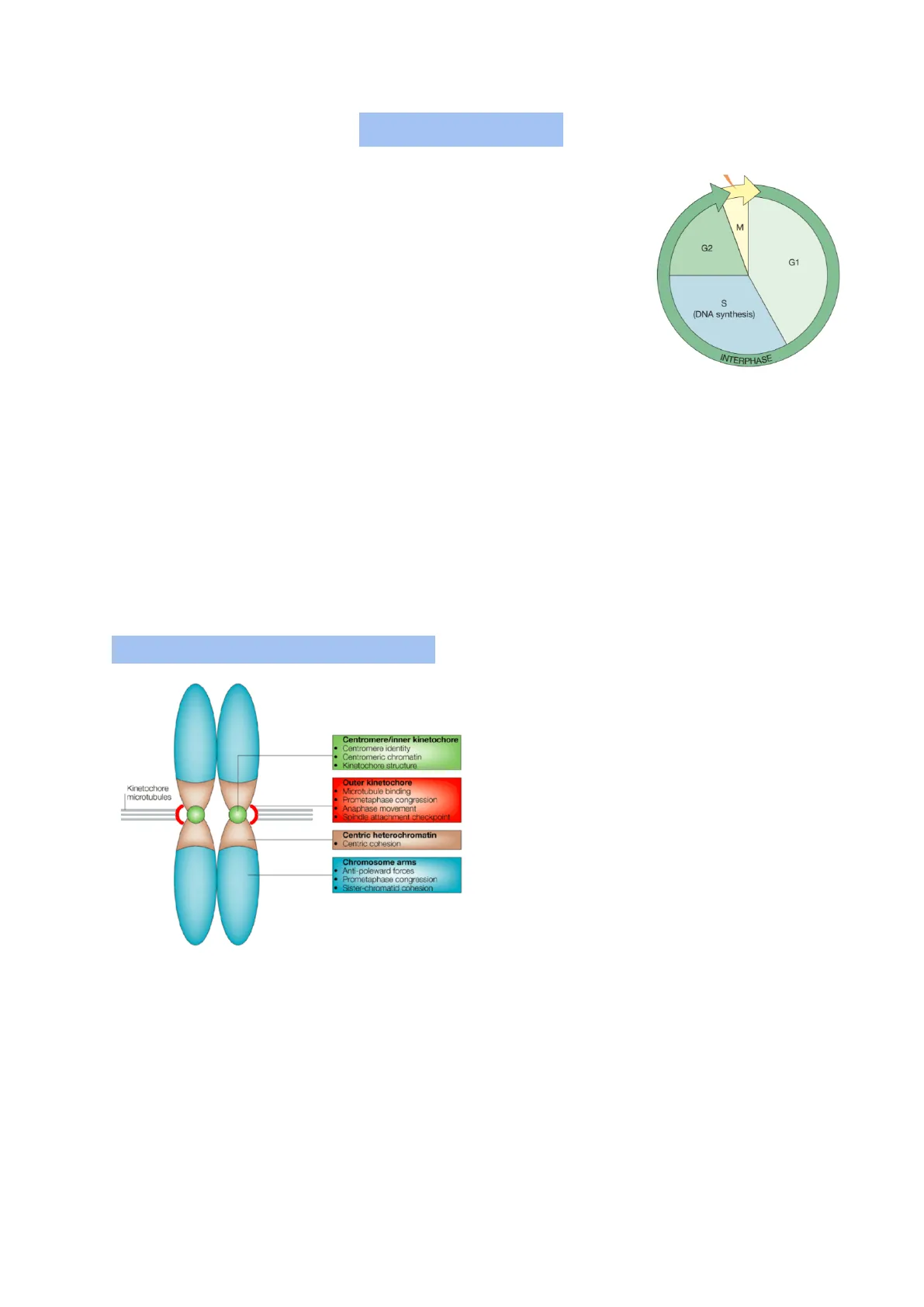
Struttura del Cromosoma
Chromosome's structure: Centromere/inner kinetochore
- Centromere identity
- Centromeric chromatin
- Kinetochore structure Outer kinetochore Kinetochore microtubules Microtubule binding Prometaphase congression Anaphase movement Spindle attachment checkpoint
- Centric heterochromatin Centric cohesion Chromosome arms Anti-poleward forces
- Prometaphase congression Sister-chromatid cohesion protein A, or CENP-A.
The centromere, marked by a primary constriction on mitotic chromosomes, contains highly repetitive DNA sequences that act as binding sites for specific proteins.
At the outer surface of the centromere on each chromatid, a protein-rich structure called the kinetochore forms. During early prophase, most of the 100-plus proteins that make up the kinetochore assemble at the kinetochore, guided by nucleosomes containing the histone variant centromeric The kinetochore functions as:
- the site of attachment of the chromosome to the dynamic microtubules of the mitotic spindle
- the residence of several motor proteins involved in chromosome motility
- a key component in the signaling pathway of an important mitotic checkpoint
M G2 G1 S (DNA synthesis) INTERPHASE 129MITOSIS
Mitosi e Citocinesi
Mitosis is a process of nuclear division in which the replicated DNA molecules of each chromosome are faithfully segregated into two nuclei. It is usually accompanied by cytokinesis, a process by which a dividing cell splits into two, partitioning the cytoplasm into two cellular packages.
The two daughter cells resulting from mitosis and cytokinesis possess genetic content identical to each other and to the mother cell from which they arose. The process maintains the chromosome number and generates new cells for the growth and maintenance of an organism. Mitosis can take place in either haploid (e.g. fungi, plant gametophytes, a few animals) or diploid cells. It is the process of division made by somatic cells.
This phase of cell cycle is completely focused on chromosome segregation. As a result, most cellular metabolic activities, such as transcription and translation, are significantly reduced, and the cell becomes less responsive to external signals.
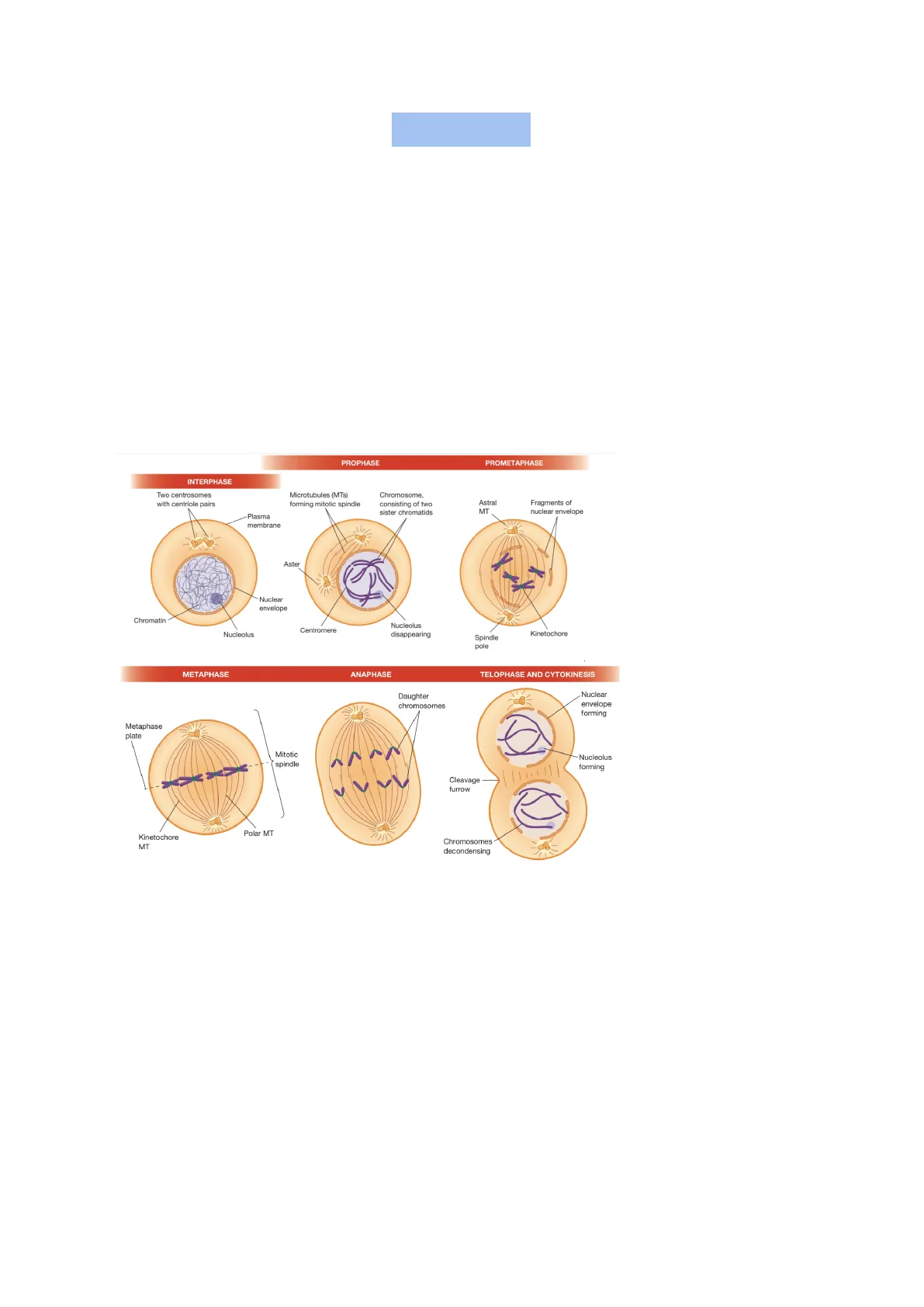
PROPHASE PROMETAPHASE INTERPHASE Two centrosomes with centriole pairs Microtubules (MTs) forming mitotic spindle Chromosome, consisting of two sister chromatids Astral MT Fragments of nuclear envelope Plasma membrane Aster Nuclear envelope Chromatin Nucleolus Centromere Nucleolus disappearing Kinetochore Spindle pole METAPHASE ANAPHASE TELOPHASE AND CYTOKINESIS Daughter chromosomes Nuclear envelope forming Nucleolus forming Cleavage furrow Polar MT Kinetochore MT Chromosomes decondensing It is divided in:
Fasi della Mitosi
Prophase:
- Chromosomal material condenses to form compact mitotic chromosomes. Chromosomes are seen to be composed of two chromatids attached together at the centromere
- Cytoskeleton is disassembled, and mitotic spindle is assembled
- Golgi complex and endoplasmic reticulum fragment.
- Nuclear envelope disperses
The nucleus of an interphase cell contains long and intertwined chromatin fibers. This state is not suited for segregation and the cell, during early prophase, converts them into much shorter, thicker structures by the process of chromosome compaction (or condensation).
The structure of mitotic and interphase chromosomes is similar (30nm in diameter).
130 Metaphase plate Mitotic spindleChromosome compaction does not alter the nature of the chromatin fiber, but rather the way that it is packaged.
Condensin e Cohesin
Condensin [non c'è nelle slide] During the process of chromosome compaction there's abundance of a multiprotein complex called condensin.
Condensin is an ATP-dependent molecular motor that translocates along double-stranded DNA, functioning as a DNA loop-extruding machine. By forming loops and introducing positive supercoiling, it compacts DNA into a smaller volume.
At the onset of mitosis, condensin is activated through phosphorylation by cyclin-Cdk complexes, making it a key target through which Cdks can trigger cell cycle activities, such as the progression from G2 to mitosis.
Cohesin During prophase, most of the cohesin is removed from the arms of the chromosomes as they become compacted. Dissociation is induced by phosphorylation of cohesin-associated proteins by two important mitotic enzymes called Polo-like kinase and Aurora B kinase.
The associated cohesins are subsequently removed from chromosome arms through the action of the protein WAPL.
Cohesin Cohesins along chromosome arms are released Cohesin remains at the centromeres because of the presence there of a phosphatase that removes any phosphate groups added to the protein by the kinases.
Centromere f region Release of cohesin from the centromeres is normally delayed until anaphase and is Cohesin remains at Chromatid centromere mediated by the protein separase.
End of S phase (uncondensed sister chromatids, arms are cohered)
Beginning of prophase (condensed sister chromatids, arms are cohered)
Middle of prophase (condensed sister chromatids, arms are free)
Formazione del Fuso Mitotico
Formation of the mitotic spindle As a cell progresses through the cell cycle, from G2 into mitosis, the microtubules of the cytoskeleton undergo rapid disassembly, preparing for their reassembly into the mitotic spindle. This disassembly is driven by the inactivation of proteins that stabilize microtubules (e.g., microtubule-associated proteins, or MAPs) and the activation of proteins that destabilize them. This process ensures that the microtubules can reorganize effectively to support chromosome segregation.
Parallel to the cell cycle, the centrosome cycle plays a key role in spindle formation. After mitosis, each daughter cell contains a single centrosome with two centrioles situated at right angles to one another. Before cytokinesis concludes, the centrioles of each centrosome disengage, losing their association with one another. This process is triggered by the enzyme separase, which becomes active in anaphase and cleaves the protein link holding the centrioles together.
131As the cell enters the S phase and DNA replication begins, each centriole starts to replicate, forming a procentriole next to the original (maternal) centriole. Microtubule elongation transforms the procentriole into a full-length daughter centriole. The centrosome duplication process begins at the G1-S transition and is triggered by the phosphorylation of a centrosomal protein by Cdk2. This tightly controlled process ensures that each mother centriole produces only one daughter centriole per cell cycle. Abnormal centrosome duplication, which can lead to extra centrioles, may result in improper cell division and potentially contribute to cancer development.
At the start of mitosis, the centrosome splits into two, each containing one pair of mother-daughter centrioles. The formation of the mitotic spindle begins in early prophase with the appearance of microtubules arranged in a "sunburst" pattern, called asters, around each centrosome. Microtubules grow by adding subunits to their plus ends, while their minus ends remain anchored at the pericentriolar material (PCM) of the centrosome. Phosphorylation of PCM proteins by Polo-like kinase is crucial for initiating the nucleation of spindle microtubules during prophase.
The formation of asters is followed by the separation of the centrosomes, which move to opposite ends of the cell. This separation is driven by motor proteins associated with adjacent microtubules. As the centrosomes separate, the microtubules between them increase in number and length, eventually positioning the centrosomes at opposite poles. This establishes the two poles of the bipolar mitotic spindle, which is essential for proper chromosome segregation. After mitosis, one centrosome is distributed to each daughter cell, ensuring continuity of the centrosome cycle.
Astral spindle microtubules Astral spindle microtubules Centriole Chromosomes Centriole Pericentriolar material Chromosomal (kinetochore) spindle fibers Polar spindle microtubules Pericentriolar material
Dissoluzione dell'Involucro Nucleare
The Dissolution of the Nuclear Envelope and Partitioning of Cytoplasmic Organelles In most eukaryotic cells, the mitotic spindle is assembled in the cytoplasm, while chromosomes remain compacted in the nucleoplasm. The interaction between the spindle and chromosomes occurs after the breakdown of the nuclear envelope at the end of prophase.
132This breakdown involves the disassembly of the three major components of the nuclear envelope:
- nuclear pore complexes: disassemble when interactions between nucleoporin subcomplexes are disrupted, causing dissociation into the surrounding medium
- nuclear lamina: breaks down due to depolymerization of the lamin filaments
- nuclear membranes: disrupted mechanically as holes are torn into the nuclear envelope by cytoplasmic dynein molecules associated with the outer nuclear membrane
The fate of the membranous portion of the nuclear envelope is debated: one view suggests the membranes fragment into vesicles that disperse, while another suggests they may be absorbed into the endoplasmic reticulum (ER).
All of these processes are initiated by phosphorylation of key substrates by mitotic kinases, particularly cyclin B-Cdk1.
Some membranous organelles, like mitochondria, lysosomes, peroxisomes, and plant chloroplasts, remain mostly intact during mitosis. The Golgi complex, which stays intact in plants, yeast, and flies, fragments in mammalian cells into vesicles and tubules, which are distributed between daughter cells and reform.
The ER network remodels into extended membrane sheets that localize to the centrosomes during metaphase, ensuring even distribution of the ER into the daughter cells.
Prometafase
Prometaphase:
- Chromosomal microtubules attach to kinetochores of chromosomes
- Chromosomes are moved to the spindle equator
The dissolution of the nuclear envelope marks the start of prometaphase. During this phase mitotic spindle assembly is completed, and chromosomes are moved at the center of cell. Initially, compacted chromosomes are scattered throughout the space that was the nuclear region. As spindle microtubules penetrate into the central region of the cell, their free (plus) ends grow and shrink in a dynamic fashion, seemingly "searching" for a chromosome. Microtubules that contact a kinetochore are "captured" and stabilized.
- A kinetochore typically makes initial contact with the sidewall of a microtubule rather than its end.
- Some chromosomes move actively along the wall of the microtubule, powered by motor proteins located in the kinetochore. Then, the kinetochore becomes stably associated with the plus end of one or more spindle microtubules from one of the spindle poles.
- A chromosome that is attached to microtubules from only one spindle pole represents an unstable intermediate stage.
4 Syntelic attachment (no tension) Tension applied by microtubules 3a 3 Bi-oriented chromosome (under tension) 1 2 Mono-oriented chromosome (no tension) (b) 133